Synthetic biology on ordinary paper: a new operating system
October 24, 2014
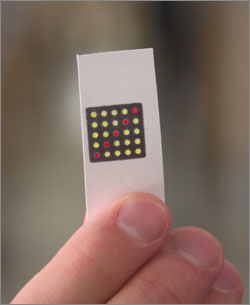
Wyss Institute scientists have embedded effective synthetic gene networks in pocket-sized slips of paper. An array of RNA-activated sensors uses visible color changing proteins to indicate presence of a targeted RNA, capable of identifying pathogens such as antibiotic-resistant bacteria and strain-specific Ebola virus. (Credit: Harvard’s Wyss Institute)
Wyss Institute for Biologically Inspired Engineering announced Thursday (Oct. 23) a way to allow complex cellular recognition reactions to proceed outside of living cells, using pocket-sized slips of paper.
Imagine inexpensive, shippable, and accurate test kits using a pocket-sized paper diagnostic tool using saliva or a drop of blood to identify specific disease or infection — a feat that could be accomplished anywhere in the world, within minutes and without laboratory support.
Two papers published Thursday in Cell describe the advances by two teams at the Wyss Institute headed by Wyss Core Faculty Members James Collins, Ph.D., and Peng Yin, Ph.D..
A new OS for synthetic biology
“What we have been able to do is to create an in vitro, sterile, abiotic operating system upon which we can rationally design synthetic, biological mechanisms to carry out specific functions,” said Collins, senior author of the first study, “Paper-Based Synthetic Gene Networks,” who is also Professor of Biomedical Engineering and Medicine at Boston University, and Co-Director and Co-Founder of the Center of Synthetic Biology.
“In the last fifteen years, there have been exciting advances in synthetic biology,” said Collins. “But until now, researchers have been limited in their progress due to the complexity of biological systems and the challenges faced when trying to re-purpose them. Synthetic biology has been confined to the laboratory, operating within living cells or in liquid-solution test tubes.”
The conventional process can be thought of through an analogy to computer programming, he says. Synthetic gene networks are built to carry out functions, similar to software applications, within a living cell or in a liquid solution, which is considered the “operating system.”
Leveraging an innovation for chemistry-based paper diagnostics previously devised by Wyss Institute Core Faculty Member George Whitesides, Ph.D. , the new in vitro operating system is ordinary paper.
“We’ve harnessed the genetic machinery of cells and embedded them in the fiber matrix of paper, which can then be freeze dried for storage and transport — we can now take synthetic biology out of the lab and use it anywhere to better understand our health and the environment,” said lead author and Wyss Staff Scientist Keith Pardee, Ph.D.
Biological programs on paper; an ebola sensor
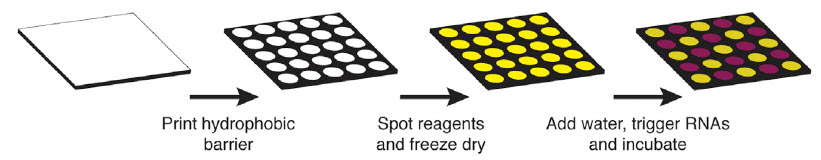
Schematic describing the process of arraying synthetic gene networks on paper using printed arrays (credit: Keith Pardee/Cell)
Using standard equipment at his lab bench and commercially-available, cell-free systems, Pardee designed and built a wide range of paper-based diagnostics and biosensors. He also used commonly-used fluorescent and color-changing proteins to provide visible indication that the mechanisms were working.
Once built, the paper-based tools can be freeze-dried for safe room-temperature storage and shipping, maintaining their effectiveness for up to one year. To be activated, the freeze-dried paper need simply be rehydrated with water.
KurzweilAI email interview with Keith Pardee
When might the first paper-based diagnostic be commercialized using the new freeze-dry method for synthetic bio parts?
We are still at pretty early stages in the development of a fieldable product. Sensitivity is our next challenge. We currently have detection into the high pico molar range, but to make the system practical for field applications, this will have to be improved. We have a number of technologies under development that promise to provide the boost to sensitivity that we need, and anticipate that we’ll reach this goal within the next 12 months.
How soon can scientists and researchers start making use of the new method in their own labs?
With the protocols we’ve included in the article, researchers can immediately start to adopt this method to applications in their lab.
When might a strain-specific Ebola sensor be commercialized using the new freeze-dry method and toehold-switch regulator?
We are working on integrating our toehold sensors with other key technologies to make it practical for the field. Areas we are focused on include improving sensitivity, like I mentioned above, and a user interface so that anyone will be able to use the diagnostic.
Has there been any interest from CDC or NIH in the Ebola sensor prototype you and Alex used in your papers? Sounds important.
This is something that will be explored going forward.
The paper-based platform can also be used in the lab to save a huge amount of time and cost as compared to conventional in vivo methods of validating tools for cell-based research. “Where it would normally take two or three days to validate a tool inside of a living cell, this can be done using a synthetic biology paper-based platform in as little as 90 minutes,” Pardee said.
As proof of concept, Collins and Pardee demonstrated a variety of effective paper-based tools ranging from small-molecule and RNA actuation of genetic switches, to rapid design and construction of complex gene circuits, to programmable paper-based diagnostics that can detect antibiotic resistant bacteria and even strain-specific Ebola virus.
The Ebola sensor was created by using the paper-based method and utilized a novel gene regulator called a “toehold switch” — a new system for gene expression control with unparalleled programmability and flexibility reported in the second study in Cell. Although its inventors had designed the toehold switch to regulate genes inside living cells, its function was easily transferred to the convenience of ordinary freeze-dried paper, showcasing the true robustness of both the freeze-dried paper technique and the toehold switch.
The Ebola sensor was conceived by Wyss Institute Postdoctoral Fellow Alex Green, Ph.D., co-inventor of the toehold switch regulator and lead author of its report, after the ongoing West Africa crisis brought the deadly pathogen to global spotlight.
Due to its easy assembly and fast prototyping ability, Green was eager to test the paper-based platform as an operating system for the toehold switch, which he had initially developed for programming gene expression in living cells. Green reached out to Pardee and together they assembled the prototype Ebola sensor in less than a day and then developed an assay that can differentiate between Sudan and Zaire virus strains within an hour of exposure.
A programmable synthetic gene regulator
The toehold switch works as such an accurate biosensor because it can be programmed to only react with specific, intended targets, producing true “switch” behavior with an unprecedented ability to turn on targeted gene expression. It can be programmed to precisely detect an RNA signature of virtually any kind and then turn on production of a specific protein.
Reported in the paper “Toehold Switches: De-Novo-Designed Regulators of Gene Expression,” Green developed the toehold switch gene regulator with senior author Yin, who is Associate Professor in the Department of Systems Biology at Harvard Medical School in addition to being a Wyss Core Faculty Member.
“While conventional synthetic biology complicates accuracy and functionality because it relies on re-purposing and re-wiring existing biological parts, the toehold switch is inspired by Nature but is an entirely novel, de-novo-designed gene expression regulator,” said Yin.
“We looked at our progress to rationally design dynamic DNA nanodevices in test tubes and applied that same fundamental principle to solve problems in synthetic biology,” said Yin. The resulting toehold switch, an RNA-based organic nanodevice, is a truly “synthetic” synthetic gene regulator with 40-fold better ability to control gene expression than conventional regulators.
The toehold switch functions so precisely that many different toehold switches can operate simultaneously in the same cell. This allows several toehold switches to be linked together, creating a complex circuit, which could be programmed to carry out multiple-step functions such as first detecting a pathogen and then delivering an appropriate therapy.
“Instead of re-purposing an existing part that was evolved by Nature, we wanted to change our way of thinking, leverage naturally-occurring principles, and build from scratch,” Green said.
By combining forces, the two Wyss Institute teams showed that the toehold switch, so effective in living cells for its dynamic control of in vivo gene expression, is also fully capable of functioning in vitro on freeze-dried paper. With its impressive gene regulation functions able to be transported out of the lab for easy delivery of diagnostics and gene therapies, paper-based toehold switches promise a profound impact on human and environmental health, the researchers say.
Powerful, multiplex biological circuits and sensors
Standing on their own, both paper-based synthetic gene networks and toehold switch gene regulators could each have revolutionary impacts on synthetic biology: the former brings synthetic biology out of the traditional confinement of a living cell, the latter provides a rational design framework to enable de-novo design of both the parts and the network of gene regulation.
But combining the two technologies together could truly set the stage for powerful, multiplex biological circuits and sensors that can be quickly and inexpensively assembled for transport and use anywhere in the world, the researchers suggest.
Harvard’s Wyss Institute | Wyss Institute scientists discuss the collaborative environment and team effort that led to two breakthroughs in synthetic biology that can either stand alone as distinct advances or combine forces to create truly tantalizing potentials in diagnostics and gene therapies.
Harvard’s Wyss Institute | In this animation, Wyss Institute Postdoctoral Fellow Alex Green, Ph.D., the lead author of “Toehold Switches: De-Novo-Designed Regulators of Gene Expression,” narrates a step-by-step guide to the mechanism of the synthetic toehold switch gene regulator.
Abstract of Paper-Based Synthetic Gene Networks
Synthetic gene networks have wide-ranging uses in reprogramming and rewiring organisms. To date, there has not been a way to harness the vast potential of these networks beyond the constraints of a laboratory or in vivo environment. Here, we present an in vitro paper-based platform that provides an alternate, versatile venue for synthetic biologists to operate and a much-needed medium for the safe deployment of engineered gene circuits beyond the lab. Commercially available cell-free systems are freeze dried onto paper, enabling the inexpensive, sterile, and abiotic distribution of synthetic-biology-based technologies for the clinic, global health, industry, research, and education. For field use, we create circuits with colorimetric outputs for detection by eye and fabricate a low-cost, electronic optical interface. We demonstrate this technology with small-molecule and RNA actuation of genetic switches, rapid prototyping of complex gene circuits, and programmable in vitro diagnostics, including glucose sensors and strain-specific Ebola virus sensors.