Animated 3D movies show cells in action over time
October 9, 2014
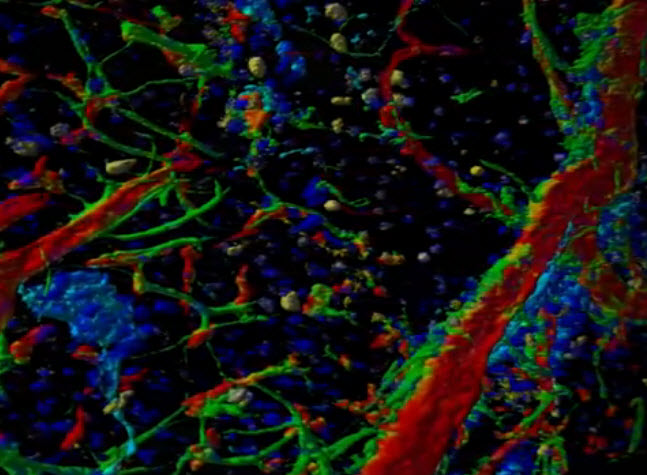
Portion of a screen from LEVER 3-D, which can produce a 3D rendering of confocal fluorescence microscopy images (shown here: mouse neural stem cells, ependymal cells, and vasculature), animated through time to show their multiplication and movement (credit: Drexel University)
Drexel University researchers have developed software and hardware that will enable biologists to use 3D glasses in a home-theater-like lab to better track and study the movement and multiplication of cells.
Their goal is to enhance the current visual data that these scientists are working with so that it’s easier to identify changes in cells over time — information that is key to studying the abnormal cell proliferation that causes cancer or the use of stem cells in regenerative medicine.
“This type of imaging is so important because it allows us to see and measure relationships between cells and their environment,” said Andrew Cohen, PhD, an associate professor of electrical and computer engineering in the College of Engineering, who is leading the group.
Cohen, whose background includes developing operating systems support for computer games, has developed a program called LEVER (Lineage Editing and Validation) that can identify, tag, and track live cells by capturing patterns of motion and cell division from sequences of microscopic time-lapse images. His work was recently published in the journal BMC Bioinformatics.
‘Photoshop’ for cell biologists
Typically the process of tracking cell lineage over time requires biologists to watch the time-lapse images and note by hand when the cells multiply — creating a graphic representation of cell division over time called a “lineage tree.” It can take a researcher several tedious hours to create a tree.
The LEVER software lets the researcher work with the computer to delineate cells, color-code them and denote the exact moment of their division. LEVER is easier and more accurate than processing the data manually and also provides more data than what can be obtained by hand.
“It’s like Photoshop for cell biologists,” Cohen said. “The software outlines cells and blood vessels, keeping track of them as they’re dividing and moving around one another. This provides a wealth of information on the patterns of cell shape, motion and division. Visualization of the 3-D microscopy data together with the analysis results is a key step to measure and ultimately understand what drives these cells.”
Animated 3D movies
With an enhanced version of the program called LEVER 3-D, which compiles data from multi-layered microscopic images, Cohen’s team can produce a three-dimensional rendering of the cells and animate it through time to show their multiplication and movement. The enhanced imaging gives researchers a unique view of the interaction of stem cells with their surroundings.
By running the software on a computer with a graphics card optimized for gaming and donning a set of 3D gaming glasses, the program can take scientists inside the microscopic cross section. In a portion of Cohen’s lab modified to be a viewing theater, a special stereo-vision projector brings the three-dimensional image data to life. Users can rotate and zoom in and out of the projected image in true 3-D, giving scientists vantage points that are not possible when looking through a microscope.
“LEVER 3-D is amazing, it opens new vistas for understanding the stem cell niche,” said Sally Temple, PhD, a cell biologist at the Neural Stem Cell Institute in Rensselaer, NY who has been using Cohen’s software as part of her stem cell research since 2005.
Cohen’s goal is to make the software open-source and readily available to any scientists who can use it for their research. The team also plans to enhance the interactive capabilities of the system to increase its utility as a tool for research. The project is funded by the National Institute on Aging.
Drexler University | SVZ Montage
Abstract of Visualization and Correction of Automated Segmentation, Tracking and Lineaging from 5-D Stem Cell Image Sequences
Neural stem cells are motile and proliferative cells that undergo mitosis, dividing to produce daughter cells and ultimately generating differentiated neurons and glia. Understanding the mechanisms controlling neural stem cell proliferation and differentiation will play a key role in the emerging fields of regenerative medicine and cancer therapeutics. Stem cell studies in vitro from 2-D image data are well established. Visualizing and analyzing large three dimensional images of intact tissue is a challenging task. It becomes more difficult as the dimensionality of the image data increases to include time and additional fluorescence channels. There is a pressing need for 5-D image analysis and visualization tools to study cellular dynamics in the intact niche and to quantify the role that environmental factors play in determining cell fate.